Molecules in Motion: Computer Simulations Lead to a Better Understanding of Protein Structures
At SDSC, McCammon and his colleagues harness 10.4 teraflops of computing power to understand how molecules naturally behave inside cells and to predict how the molecules might react to the presence of prospective drugs. (One teraflop is equal to one trillion operations per second.)
By exposing a weakness in the only HIV enzyme not targeted by the antiretroviral drugs currently used to fight HIV, McCammon's models revealed a path for the development of new treatments.
Traditionally, researchers have studied the structures of enzymes and other proteins by growing protein crystals and shining x-rays at them, explains Kamala Shukla, program director for the Biomolecular Systems Cluster in the Division of Molecular and Cellular Biosciences at NSF. Interpreting the x-ray scans reveals a 3-dimensional structure of the molecule. But the method, called x-ray crystallography, doesn't allow researchers to visualize molecules in motion.
"Suppose you see me standing up in a room," Shukla explains. "At that point, you know my structure. But you don't know what I'm going to do, or how I might change over time."
It's a similar story with enzymes and other proteins.
"These protein structures were always depicted on the covers of Scientific American and journals such as Science and Nature as very static images," McCammon said in an interview published online by the Smithsonian Institution, "and so the great majority of the scientific community tended to think of proteins as fairly rigid structures."
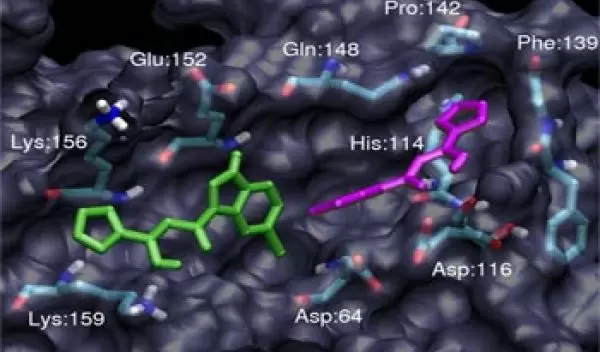
Accordingly, scientists thought of the drugs that inhibit enzymes as "keys" that could fit into the locks, or active sites, of a given enzyme. The lock-and-key model was useful but limited when it came to enzymes with especially complex structures. As computing power grew in the 1990s, researchers were able to develop more sophisticated models, and these simulations began to show that enzymes are not rigid like steel, but flexible, more like plastic.
To further complicate matters for structural biologists, enzymes are large--sometimes made of thousands of molecules--and irregularly shaped. Often, the biologically important domains--the active sites--are hidden within intricately folded sheets.
But plugging thermodynamics equations and details known from x-ray crystallography studies into computer simulations can reveal hidden domains and information about how molecules move. It is here, at the nexus of biology and computer science, where McCammon and his colleagues study how molecules might respond to new drugs before any experiments are conducted in petri dishes or on animals. The researchers are pioneering an emerging field known as bioinformatics, the use of computational tools to attack biological problems.
In 1976, when McCammon received his first NSF grant as a postdoctoral researcher, such practical applications seemed a remote possibility. "We had no idea at the time what this might be useful for, if anything. It was purely abstract research...and a good example of how something that starts out as pure research, not goal-oriented in any sense whatsoever, turns out to be tremendously useful and practical for applications later on," McCammon says.
When HIV was identified as the agent of AIDS in the early 1980s, scientists began to realize that computer simulations of HIV enzymes could help in drug discovery, McCammon says.
Like most RNA viruses (viruses that rely on RNA rather than DNA as their genetic material), HIV mutates quickly, and some new strains show resistance to even the most potent cocktails of available anti-retroviral drugs. Resistance has become a "very serious problem," McCammon says, so the need for new therapies is acute.
A 2002 study published in the New England Journal of Medicine found that the prevalence of drug-resistant strains of HIV rose from 3.4 percent of HIV-infected patients in 1995 to 12.4 percent in 2000. And according to a 2004 review article in the same publication, as many as half of all patients receiving antiretroviral therapy are infected with viruses that are resistant to at least one of the available antiretroviral drugs.
HIV relies on the activity of three enzymes to survive and proliferate in the body: reverse transcriptase, protease and integrase. Before McCammon's simulations, the structure of the third enzyme, integrase, was not understood well enough to be successfully targeted with drugs. Integrase catalyzes a critical step in the HIV life cycle, directing the viral genetic material to mix with the host cell's DNA. If integrase activity could be inhibited, the virus would have no way of hijacking the cellular machinery to replicate itself.
Researchers at Merck Pharmaceutical Company have used McCammon's work to design new drugs that target integrase, and the new compounds are now entering human trials.
If integrase inhibitors effectively treat HIV in people, it will be a great success for McCammon and his team. But they don't expect the story will end there: "We hope that the methods demonstrated in this work will help in the development of drugs for other diseases, including other infectious diseases like influenza, and diseases such as cancer and diabetes."
McCammon holds the Joseph Mayer Chair of Theoretical Chemistry at the University of California, San Diego. The San Diego Supercomputer Center was founded in 1985 with a $170 million grant from the NSF Supercomputer Centers program.
--Sarah Goforth


