News Release 05-095
Zipped Structure May Explain Protein Clumping in Brain Disorders
Finding may provide insight on Alzheimer's and Huntington’s disease
June 8, 2005
This material is available primarily for archival purposes. Telephone numbers or other contact information may be out of date; please see current contact information at media contacts.
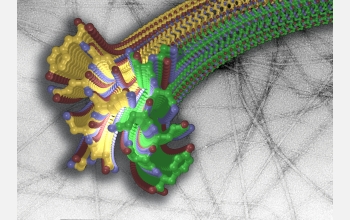
After years of intense work, researchers have discovered the 3-dimensional structure of a miniscule--yet mighty--region of a protein that forms deleterious rope-like structures in the brain. Known as amyloid fibrils, the proteins are associated with the degenerative brain disorders Alzheimer's, Parkinson's and Huntington's diseases, and so-called prion diseases like mad cow. This particular region of the protein catalyzes the formation of a "molecular zipper," which pulls proteins together to form the stubbornly stable clumps.
Knowing the structure will help researchers devise new treatments for the more than two-dozen human diseases associated with fibrils, which are attributed to killing neurons and other types of cells. Effective therapeutics may reverse the zipping to break down persistent fibrils or prevent them from forming in the first place.
The work appears in the June 9 issue of the journal Nature.
By studying a specific yeast protein that aggregates similarly to the fibrillar proteins associated with animal diseases, David Eisenberg, his team at the University of California, Los Angeles (UCLA), and international colleagues determined that a region of these fibril-forming proteins forms two sheets that "zip together." This coupling--occurring along a self-guided track--squeezes out water molecules to form a dry, persistent structure that helps account for the tenacity of fibril build ups.
This abnormally dry, zipped-up protein is completely insoluble, that is, it does not dissolve in water, which is a hallmark of amyloid fibrils. In people with Alzheimer's disease, for example, the build up of fibrils in the brain is commonly referred to as plaque.
After years of solo work on the protein's structure, Eisenberg teamed with Christian Reikel, an expert who studies the crystal structures of small-scale molecules at the European Synchrotron Radiation Facility. Using a technology that allowed them to "see" the protein's 3-dimensional structure, the combined group is the first to document the molecular structure of a fibril--a feat that has eluded researchers for decades.
This research is supported by the National Science Foundation, the National Institutes of Health, the Howard Hughes Medical Institute and the U.S. Public Health Service.
-NSF-
Media Contacts
Richard (Randy) Vines, NSF, (703) 292-7963, email: rvines@nsf.gov
Principal Investigators
David Eisenberg, University of California, Los Angeles, (310) 825-3754, email: david@mbi.ucla.edu
Related Websites
For a complete story, see the UCLA news release at: http://newsroom.ucla.edu/page.asp?RelNum=6197
The U.S. National Science Foundation propels the nation forward by advancing fundamental research in all fields of science and engineering. NSF supports research and people by providing facilities, instruments and funding to support their ingenuity and sustain the U.S. as a global leader in research and innovation. With a fiscal year 2023 budget of $9.5 billion, NSF funds reach all 50 states through grants to nearly 2,000 colleges, universities and institutions. Each year, NSF receives more than 40,000 competitive proposals and makes about 11,000 new awards. Those awards include support for cooperative research with industry, Arctic and Antarctic research and operations, and U.S. participation in international scientific efforts.
Connect with us online
NSF website: nsf.gov
NSF News: nsf.gov/news
For News Media: nsf.gov/news/newsroom
Statistics: nsf.gov/statistics/
Awards database: nsf.gov/awardsearch/
Follow us on social
Twitter: twitter.com/NSF
Facebook: facebook.com/US.NSF
Instagram: instagram.com/nsfgov