News Release 07-102
Scientists Retrace Evolution of an Important Human Protein
Study improves our understanding of evolution by identifying steps in the evolution of a protein over the last 450 million years.
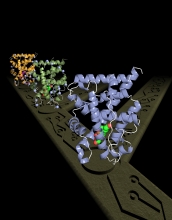
Structural evolution of an important protein in humans and other vertebrates.
August 16, 2007
This material is available primarily for archival purposes. Telephone numbers or other contact information may be out of date; please see current contact information at media contacts.
Scientists have, for the first time, identified the atomic structure of an ancient protein and revealed in unprecedented detail how it evolved into an important protein in humans and other vertebrates. In doing so, the scientists also addressed some long-standing controversies in evolutionary genetics.
"Never before have we seen so clearly, so far back in time," said Joe Thornton of the University of Oregon. "We were able to see the precise mechanisms by which evolution molded a tiny molecular machine at the atomic level, and to reconstruct the order of events by which history unfolded."
A paper describing the protein's evolution appears August 16 in Science Express, an online publication where the journal Science promotes selected research before regular publication.
A detailed understanding of the evolution of proteins--the workhorses of every cell--has long eluded evolutionary biologists, largely because ancient proteins have not been available for direct study. But in a study that was partially funded by the National Science Foundation (NSF), Thornton and Jamie Bridgham, a postdoctoral scientist, used state-of-the-art computational and molecular techniques to recreate the progenitors of an important protein known as the glucocorticoid receptor (GR).
Next, Thornton collaborated with University of North Carolina biochemists Eric Ortlund and Matthew Redinbo, who used ultra-high energy X-rays from a stadium-sized Advanced Photon Source at Argonne National Laboratory near Chicago to chart the precise position of each of the 2,000 atoms in the ancient proteins. The researchers then worked together to trace how changes in the protein's atomic architecture over the last 450 million years enabled GH to evolve its current unique function: to enable cells to respond to the hormone cortisol, which regulates the body's stress response.
"This is the ultimate level of detail," Thornton said. "We were able to see exactly how evolution tinkered with the ancient structure to produce a new function that is crucial to our own bodies today. Nobody's ever done that before."
The techniques used in this study were incredibly creative and integrative," says Dianna Padilla, an NSF program director. "These researchers have achieved something that other scientists only dream of."
Through their analyses of atomic structures, the scientists determined that just seven historical mutations were needed over the last 450 million years for GR's progenitors to evolve GR's present-day response to cortisol. The scientists also discovered that for the protein to acquire its response to cortisol, certain major mutations had to have been preceded by certain other relatively small, otherwise negligible changes, known as "permissive" changes. Such relationships helped the scientists deduce the order in which all of the gene mutations occurred.
Specifically, Thornton's studies showed that the evolving protein's most radical mutation remodeled a whole section of the protein, bringing a group of atoms close to the hormone. A second mutation in this repositioned region then created a tight new interaction with cortisol. Other earlier permissive mutations buttressed particular parts of the protein so they could tolerate this eventual remodeling.
"These permissive mutations are chance events," Thornton said. If they hadn't happened first, then the path to the new function could have become an evolutionary road not taken. Imagine if evolution could be rewound and set in motion again: a very different set of genes, functions and processes might be the outcome."
"This study has refined our knowledge of evolution because it helps address the question of whether adaptation occurs through large or small effect mutations," said Padilla. "This study shows that small changes may enable large adaptations to occur. These large adaptations may then by further refined by smaller adaptations."
Other organizations besides NSF that helped fund this project included the National Institutes of Health, the UNC Lineberger Comprehensive Cancer Center and an Alfred P. Sloan Research Fellowship to Thornton.
-NSF-
Media Contacts
Lily Whiteman, NSF, 703-292-8310, email: lwhitema@nsf.gov
Zack Barnett, University of Oregon, 541-346-3145, email: zbarnett@uoregon.edu
Program Contacts
Mary Chamberlin, NSF, (703) 292-8413, email: mchamber@nsf.gov
Principal Investigators
Joe Thornton, University of Oregon, 541-914-2588, email: joet@uoregon.edu
The U.S. National Science Foundation propels the nation forward by advancing fundamental research in all fields of science and engineering. NSF supports research and people by providing facilities, instruments and funding to support their ingenuity and sustain the U.S. as a global leader in research and innovation. With a fiscal year 2023 budget of $9.5 billion, NSF funds reach all 50 states through grants to nearly 2,000 colleges, universities and institutions. Each year, NSF receives more than 40,000 competitive proposals and makes about 11,000 new awards. Those awards include support for cooperative research with industry, Arctic and Antarctic research and operations, and U.S. participation in international scientific efforts.
Connect with us online
NSF website: nsf.gov
NSF News: nsf.gov/news
For News Media: nsf.gov/news/newsroom
Statistics: nsf.gov/statistics/
Awards database: nsf.gov/awardsearch/
Follow us on social
Twitter: twitter.com/NSF
Facebook: facebook.com/US.NSF
Instagram: instagram.com/nsfgov


