Fighting the Flu
Each year, people everywhere prepare for flu season. Some will get the flu vaccine, some take vitamin supplements, some launch a vigorous handwashing campaign, while others take an entirely different approach: nuclear magnetic resonance (NMR).
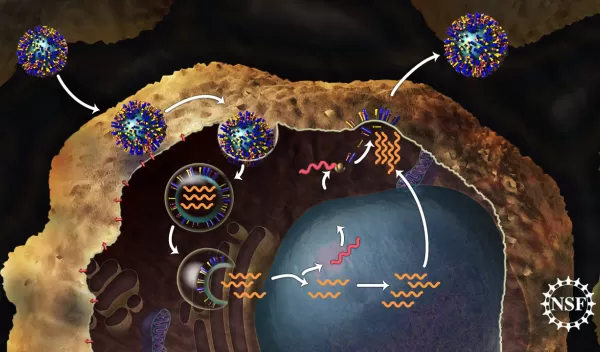
Mei Hong, a chemist at Iowa State University, uses NMR to study a proton channel named M2, found on the surface of the influenza virus. This proton channel has been the subject of numerous studies because of the key role it plays in helping a virus take over a healthy cell after it gets inside.
In order to infect a healthy cell, a flu virus must enter the cell and empty viral genes into it. First, the virus binds to the cell. Then it becomes enveloped inside a bubble called an endosome as it gets taken into the cell. It is acidic inside the endosome--more acidic than the interior of the virus it contains. This pH difference serves as a signal to the virus that it is inside the cell, so it is time to release the viral genes. It is the M2 proton channel that helps the virus sense this difference in acidity and trigger the release of viral genes into the cell. Since the cell cannot tell the difference between its own genes and those of the virus, it gets tricked into making copies of the virus which can eventually go on to infect other cells.
Another great unknown
We know quite a bit about how a virus can infect a cell, but until recently it was still unclear exactly how the protons move from the endosome into the virus to trigger infection. Scientists suggested two distinct possibilities: the first, a "shutter" model in which the channel is either "open" or "closed" to the flow of water that carries an extra proton into the virus, or a "shuttle" model, in which the components of the channel actually move to relay protons from the exterior to the interior of the virus.
"Computational methods for predicting these structures are not good enough yet, so experimentally determining the high resolution structure of the histidine portion of this molecule, which controls the proton relay, is very important. Fortunately we had a vast knowledge of this protein that allowed us to zoom in on the histidine residue precisely," said Hong.
The role of NMR
But what is NMR, and how do researchers use it? NMR is actually a property of the nucleus of an atom when it interacts with a magnetic field. What that means is that NMR is a property of matter, like color or density or mass. Scientists have found ways to turn this property into useful information, especially in the field of imaging. For example, physicians use magnetic resonance imaging (MRI) machines to see inside a patient without surgery. These machines use NMR to create a detailed visualization of tissues in the body. Scientists can use NMR in the same way on a really magnified scale to look inside much smaller things like an individual protein, or even just a part of a protein.
"One can study the structure of proteins in many ways, but protons are difficult to visualize by X-rays," said Hong. X-rays are commonly used to study the structure of proteins in a crystal form. "We used NMR because we wanted to see where the protons are relative to the channel. Moreover, the state of the samples that we study using NMR is closer to the natural biological state than most other structure determination techniques."
Shuttling through
Using NMR, Hong's team was able to obtain evidence that the channel moves protons by the "shuttle" model. The work, published in the October 22, 2010 issue of the journal Science, shows that ring-shaped histidine molecules facing the pore of the channel are able to flip back and forth at more than 50,000 times per second when the channel is in an open state (imagine a windshield wiper-like motion).
After every few flips, the histidine is able to pick up a proton from outside the virus and flip it into the interior of the virus. Movement of protons into the virus causes the interior to become more acidic as protons accumulate. This serves as a signal that the virus is inside the cell, and it should release its genes--the next step in the viral infection process.
It's a shuttle. So what?
Knowing which model is the right one is actually pretty important, because there are drugs that can block this step in the process of viral infection. Amantadine and rimantadine, respectively known by the trade names Symmetrel and Flumadine, are two of the older antiviral drugs approved by the FDA. Both have been shown to make flu symptoms less severe and shorten the time it takes to get better.
Hong has also used NMR to learn more about amantadine, which prevents susceptible viruses from spreading. The study, included in Nature on February 4, 2010, showed that amantadine binds two different sites on the M2 protein. Combined with other data, Hong and colleagues concluded that amantadine physically blocks the M2 channel, preventing the movement of protons into the virus and subsequently, reproduction and spread of the virus.
Unfortunately, amantadine and rimantadine are no longer recommended for widespread use because many flu strains, including H1N1, are resistant.
Bring on the mutants
Now that scientists have a better understanding of how the M2 channel moves protons and where amantadine binds in the M2 channel, they want to use the same techniques to study how the M2 channel is different in drug-resistant versions of the virus.
Hong explained, "Since most flu viruses now have the mutation, we will focus on that next."
Results from the Nature paper hint that amantadine did not fit well into the M2 channel. One strategy could be to design a drug that is better able to occupy the known binding sites, but now that we know the channel must "shuttle" the protons through, Hong says there are more options: "Targeting the histidine may work just as well to block the channel."
New and better antiviral drugs will aid those that didn't get vaccinated, or those that become ill before their vaccine becomes fully effective. Millions of people in the United States get the flu each year. The majority will recover without treatment, but thousands become sick enough to be hospitalized. According to CDC estimates, there are anywhere from 3,000 to 49,000 flu-related deaths each year. Effective drug treatment could prevent hospitalizations and reduce the number of complication-related deaths.
Fighting disease in the future
Research in this area will provide benefits beyond development of better antiviral drugs. Eventually, scientists will have enough data from NMR experiments to work with mathematicians on creating models that can more accurately predict the shape and structure a protein will have. These models, once tested and refined, could help researchers predict and identify previously unknown molecular interactions. A better understanding of the myriad of players that interact to make us feel good or bad each day could play a significant role in shaping future medical diagnostics and treatments. Studies that allow increasingly accurate unification of protein structure and function, like the NMR experiments done by Mei Hong, are steps on the path towards more effective, personalized medical treatments.


