Credit: National Museum of American History, Smithsonian Institution
From punch cards to protein powerhouse
When NSF first started funding the PDB in 1975, the data bank was small and local, serving specialists in structural research. The inventory had launched in 1971 at Brookhaven National Laboratory and contained the 3D structures for seven molecules. These were shared with requestors by mail using either magnetic tape or punch cards, with each card containing data for a single atom.
NSF's support, joined by the U.S. Department of Energy and the National Institutes of Health in 1989, dramatically boosted the repository's reach and size. By 2003, the groups supporting the PDB had gone global, with a consortium of universities, the National Institute of Standards and Technology and European and Japanese institutes forming the Worldwide Protein Data Bank (wwPDB).
From its modest origins, the PDB has grown into a powerful digital resource, housing over 230,000 structures and serving researchers from across scientific disciplines, the pharmaceutical and biotechnology industries, and educators and students from all levels.
The PDB's first molecules were simple ones whose structures had been determined by a method known as X-ray crystallography, where diffracted X-rays generate a pattern that reveals an atom's structural orientation.
Later, more powerful techniques like nuclear magnetic resonance, which uses the local conformation and distance between atoms, and electron microscopy, which uses images of molecular shapes, helped the PDB grow, adding larger, more complex molecules to its inventory.
Structuring discoveries
With over 60,000 contributors globally and millions of annual users, the PDB has revolutionized drug development, informed patentable technologies and enabled new methods for scientific discovery. Some pioneering breakthroughs include:

From ion channels to targeted therapeutics
Ion channels transmit electrical signals between cells and are essential for movement and sensation.
NSF-funded researcher Roderick MacKinnon's molecular description of ion channels, awarded the 2003 Nobel Prize in chemistry, advanced the understanding of cell biology, such as mechanisms that control heartbeat and brain function, leading to targeted therapeutics for epilepsy, cystic fibrosis and cardiovascular diseases.
From ribosomes to better antibiotics
Ribosomes are the cell's protein factories, producing the proteins essential for all organisms.
In 2000, Venkatraman Ramakrishnan, Ada E. Yonath and NSF-funded researcher Thomas A. Steitz provided the first ribosomal structures. This work advanced the understanding of ribosomal function and improved the efficacy of ribosome-targeting antibiotics, earning them the 2009 Nobel Prize in chemistry.

From photosynthesis to biotechnology
Photosystem I is a protein complex that enables photosynthesis — the conversion of light to chemical energy — an essential process for much of life.
NSF-supported researchers improved structural determination of photosystem I, paving the way for living fuel cells that harness photosynthesis to generate electricity.
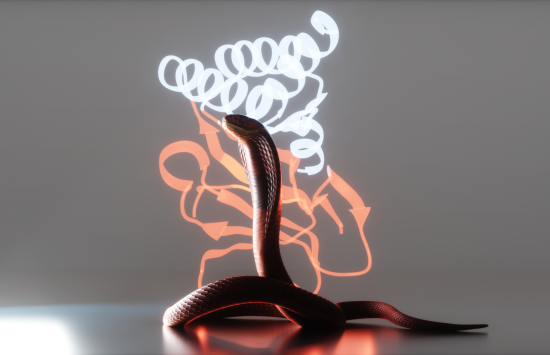
From protein design to antitoxins
PDB data were critical for computational protein design by NSF-funded researcher David Baker and AI-powered protein structure prediction tool AlphaFold2, led by Demis Hassabis and John Jumper.
These breakthroughs, which earned the three researchers the 2024 Nobel Prize in chemistry, have accelerated research to improve medical treatments, including new proteins that neutralize toxins from deadly cobras.
A massive repository at the tip of your fingers
What has made the PDB as big as it is today? From its early days, the data bank has been a model for the open-access movement, inspiring similar repositories in fields like genomics and neuroscience. Many academic journals require that researchers publish molecular structures in the PDB as a condition for publication, ensuring that people across the globe have access to this essential data.
In addition to hosting molecular structures on the PDB site, the activities, videos and multimedia found on its PDB 101 portal are used by nearly 1 million educators, students and members of the public each year to learn about the molecules of life.