Multinational Coordinated Arabidopsis thaliana Functional Genomics Project: Beyond the Whole Genome Sequence: June 2002
 On this page
On this page
Members of the Multinational Arabidopsis Steering Committee
Michael Sussman, Co-chair, University of Wisconsin, representing the United States
Thomas Altmann, Co-chair, University of Potsdam / Max-Planck- Institute of Molecular Plant Physiology, representing Germany
Bill Crosby, NCR Canada, representing Canada
Ian Furner, University of Cambridge, representing the United Kingdom
Mary Lou Guerinot, Dartmouth College, representing the United States
Gerd Jürgens, University of Tübingen, representing Germany
Ottoline Leyser, University of York, representing the United Kingdom
Jiayang Li, Chinese Academy of Sciences, representing China
Keith Lindsey, University of Durham, representing the United Kingdom
Sean May, Nottingham Arabidopsis Stock Center, representing the United Kingdom
Peter McCourt, University of Toronto, representing Canada
Fernando Migliaccio, Consiglio Nazionale delle Ricerche (CNR), representing Italy
Javier Paz-Ares, Centro National de Biotechnoligia, representing Spain
Kazuo Shinozaki, RIKEN Tsukuba Institute, representing Japan
Ian Small, INRA-Evry, representing France
Geoff Wasteneys, Australian National University, representing Australia and New Zealand
ZhangCai Yan, NSFC, representing China
Marc Zabeau, European Plant Sciences Organization
Rebecca Joy, Executive Secretary and Coordinator
Foreword to the Report
In 1990, an ad hoc committee composed of nine scientists from the United States, Europe, Japan and Australia prepared a report called “Long-range Plan for the Multinational Coordinated Arabidopsis thaliana Genome Research Project.” The report outlined a plan for international cooperation in studies of the model plant, Arabidopsis thaliana. The mission statement read “The mission of the project is to identify all of the genes by using a functional biological approach leading to determination of the complete sequence of the Arabidopsis genome by the end of this century.” The stated ultimate goal of the project was “to understand the physiology, biochemistry, and growth and developmental processes of a flowering plant at the molecular level, using Arabidopsis as an experimental model system.”
At the end of the 20th Century, the complete genome sequence of Arabidopsis was published, thus accomplishing the mission of the Multinational Coordinated Arabidopsis thaliana Genome Research Project. Analysis of the complete genome sequence indicates that there are approximately 25,500 genes in Arabidopsis. Now the Arabidopsis research community is proposing a new mission: to determine the function of every gene in Arabidopsis by 2010. The ultimate goal remains the same: a complete understanding of the biology of a flowering plant, using Arabidopsis as an experimental model system.
The purpose of this document is to outline what is required in the next ten years for the Arabidopsis research community to accomplish the new mission. The availability of the complete genome sequence gives us for the first time a glimpse of the information needed for a complete understanding of plant biology. However, like the charcoal sketches a painter draws on a canvas, this sequence is just a framework: an outline that provides data, but little understanding. Exploitation of this information will establish Arabidopsis as the premier species for the complete study of the physiology, biochemistry, and development of plants, and will serve as a basis for comparative studies as well as application of the knowledge gained to more economically important species.
Secondly, this document will serve as an update to the Arabidopsis research community at large of the efforts being made in Arabidopsis Functional Genomics world-wide. To maintain optimal research efficiency, it is important to keep the community current not only on the status of research but on the funding, biological resources and services being made available around the world that will drive this research forward.
It has became clear during the past ten years that international cooperation and communication are essential elements to success in an undertaking as large as the Multinational Coordinated Arabidopsis thaliana Functional Genomics Project. We have put forth a goal of no less than complete understanding of the biology of an organism; the only way to achieve success is to work together with the realization that we are all wedded to the same goal.
The Multinational Arabidopsis Steering Committee
June 2002
Executive Summary
A long-range plan for the Multinational Coordinated Arabidopsis thaliana Functional Genomics Project has been developed by the international community of scientists engaged in the study of basic plant biology using Arabidopsis as a model system. The project was conceived and developed in response to the completion of the Arabidopsis genome sequence by the Arabidopsis Genome Initiative in December of 2000. For the first time, scientists have access to the sequence of the 25,500 genes required for the functioning of a flowering plant. This information brings with it an opportunity: the chance to exploit this new-found knowledge to bring about the complete understanding of plant biology.
The international community of plant scientists enters into the new era of functional genomics research with the realization that in order to meet the needs of an expanding world population and of protecting the environment for future generations, we must find ways to improve the plants that we rely on for our existence; that before we can efficiently make improvements to economically important plant species, we must further our knowledge of plant biology; and that the best way to rapidly and efficiently gain this knowledge is through the use of the experimental model system Arabidopsis thaliana.
Mission: To determine the function of every gene of a reference species in its cellular, organismal, and evolutionary context by the year 2010.
Project Goal: The ultimate goal of the project is a complete understanding of the biology of a flowering plant, using Arabidopsis as an experimental model system.
Scientific Objectives:
- Development of an expanded genetic toolkit, including new technology development that enables a broad community of scientists to conduct functional genomics research in Arabidopsis
- Whole-systems identification of gene function, including global analyses of gene expression, the plant proteome, metabolite dynamics, molecular interactions and comparative genomics
- Expansion of the role for bioinformatics
- Development of community and human resources
- Promotion of international cooperation
International Collaboration: It is recognized by the community of Arabidopsis researchers that the success of the Multinational Coordinated Arabidopsis thaliana Genome Research Project was due in large part to the tremendous amount of international cooperation that was an integral part of the project. As we launch this new project, we take with us this lesson: international cooperation, coordination of efforts, and communication among the involved groups are essential. The Multinational Arabidopsis Steering Committee will continue to provide the means for ensuring a high level of international cooperation.
Introduction
Humans depend on plants in nearly every aspect of life. We use plants for food, both directly and as secondary consumers. We use plant structural components as building materials and textiles, and plant metabolites for their nutritional and medicinal properties and as industrial raw materials. Photosynthesis provides the biological and chemical energy that fuels our world and is responsible for the oxygen and carbon dioxide cycling that makes our very existence on Earth possible. The importance of plants in our world cannot be overemphasized.
Although these facts have been true throughout human history, gaining knowledge about the biology of plants has never been more important than at this moment. The population of the world is expanding rapidly: at 2.5 billion in 1950, the population has more than doubled to 6.2 billion today, and is estimated to reach the 7 billion mark in just over ten years (US Census Bureau). It seems clear that in order to feed this growing population, world food production must be increased, especially in regions of the world with the greatest population density. Because the world’s arable land is already utilized almost to its limit, it will be necessary to find new ways to improve crop yields, and to do so in an environmentally friendly fashion.
Large strides have been made in plant research in the last decade. Research has given us insight into the natural processes of disease resistance, response to environmental stresses and plant metabolism, to name just a few. We have also begun to understand the developmental processes, biochemistry, and physiology of many species of plants.
This research has provided tantalizing clues about a future in which we can utilize our knowledge to make positive changes in plant species of economical importance. Possibilities include enhancing resistance to disease caused by insect, bacterial, viral and parasitic pests; increasing tolerance to abiotic stresses, such as heat, drought and soil salinity; and doing these things while decreasing dependence on chemical fertilizers and pesticides. The amount of an important nutrient that is present at unhealthy or low levels in a crop can be increased, other nutrients introduced, or adverse components removed to enhance nutritional value. The net result will be plants that produce more, higher quality food and resources for our growing population.
These goals cannot be accomplished until we have achieved a deep and thorough understanding of plant biology. We cannot hope to improve a plant until we know how it functions under normal conditions, how it responds to altered conditions, and how such a response affects the physiology of the entire organism. The task of achieving an exhaustive knowledge of the biology of even one plant seems daunting, indeed. However, it must be done.
As all properties of a living organism are determined by its genetic constitution through interaction with its environment, the starting point is to discover the structure and function of each gene of a flowering plant and determine its role in the control of the metabolic and developmental processes of the plant.
Arabidopsis: The Model Plant
Arabidopsis thaliana is a small dicotyledonous species, a member of the Brassicaceae or mustard family. Although closely related to such economically important crop plants as turnip, cabbage, broccoli, and canola, Arabidopsis is not an economically important plant. Despite this, it has been the focus of intense genetic, biochemical and physiological study for over 40 years because of several traits that make it very desirable for laboratory study. As a photosynthetic organism, Arabidopsis requires only light, air, water and a few minerals to complete its life cycle. It has a fast life cycle, produces numerous self progeny, has very limited space requirements, and is easily grown in a greenhouse or indoor growth chamber. It possesses a relatively small, genetically tractable genome that can be manipulated through genetic engineering more easily and rapidly than any other plant genome.
Arabidopsis, like all flowering plants, dehydrates and stores its progeny at ambient temperature for long periods of time. This fact, together with a newly developed means of creating gene knockout lines, has made many basic biologists realize that Arabidopsis may be the best model system for basic research in the biology of all multicellular eukaryotes. A complete knockout collection of Arabidopsis seeds can be housed in a room no larger than a closet (see photo); to create and store a similar library of knockouts for mouse, flies and worms would be much more laborand space intensive. All together, these traits make Arabidopsis an ideal model organism for biological research and the species of choice for a large and growing community of scientists studying complex, advanced multicellular organisms.Arabidopsis versus plants of economic significance
Why Arabidopsis? Why not concentrate our research efforts and resources on a species that will actually provide food for our world or useful products for industrial uses? In order to make the strides necessary to increase crop production in a relatively short time, we have to be able to move forward quickly and spend the available human and financial resources as efficiently as possible. This is the advantage of a model system: an organism that is easily manipulated, genetically tractable, and about which much is already known. By studying the biology of Arabidopsis, the model plant, we can gain comprehensive knowledge of a complete plant. In the laboratory, Arabidopsis offers the ability to test hypotheses quickly and efficiently. With the knowledge we gain from the model plant thus established as a reference system, we can move forward with research and rapidly initiate improvements in plants of economic and cultural importance.
One advantage offered to the plant researcher by Arabidopsis is its relatively small genome size. Many crop species have large genomes, often as a result of polyploidization events and accumulation of non-coding sequences during their evolution. Maize has a genome of approximately 2400 Megabase pairs (Mbp) – around 19 times the size of the Arabidopsis genome – with probably no more than double the number of genes, most of which occur in duplicate within the genome. The wheat genome is 16000 Mbp – 128 times larger than Arabidopsis and 5 times larger than Homo sapiens – and it has three copies of many of its genes. The large crop genomes pose challenges to the researcher, including difficulty in sequencing as well as in isolation and cloning of mutant loci.
Evidence from the rice genome project suggests that the Arabidopsis genome may be missing some homologs of genes present in the rice genome. Despite this, most of the difference in gene number between Arabidopsis and crop species appears to result from polyploidy of crop species’ genomes, rather than from large classes of genes present in crop species that are not present in Arabidopsis. Therefore, the genes present in Arabidopsis represent a reasonable model for the plant kingdom. However, it is clear that Arabidopsis represents a starting point rather than the finish line for utilizing the full power of genomics for crop improvement.A tradition of Arabidopsis research
Arabidopsis has been the organism of choice for many plant biochemists, physiologists, developmental biologists and geneticists for several decades. In that time, a great deal of knowledge has been gained about the biology of this flowering plant. With the completion of the Arabidopsis genome sequencing project, we now have in hand the sequence of the approximately 25,500 genes in its genome. An extensive toolkit for manipulation has been developed over the last 20 years, including efficient mutagenesis, facile transformation technology, and DNA, RNA, protein, and metabolite isolation and detection methods. The biological reagents that have been made available to the community enable rapid research progress. Ongoing research within the community has resulted in working knowledge of many of the biochemical, physiological, and developmental processes of Arabidopsis.
Technological innovation and education
The availability of a broad base of knowledge about Arabidopsis and the previously developed research toolkit invites scientists to establish new techniques, develop new approaches, and test new concepts in Arabidopsis prior to their application in other species. The novel technologies made available in this way not only continually increase the efficiency of research done in Arabidopsis, but expose researchers, most importantly young scientists, to the most up-to-date methods in plant research, which they can apply to other species as they move forward in their career.
Arabidopsis research is the first step in an exciting future of plant improvement
Much work remains to be done before the goal of complete knowledge of the biology of even one plant species comes to fruition. It is essential that the work leading to the achievement of this goal be done as quickly and efficiently as possible. When we have achieved this ambitious goal, we will have the power to predict experimental results and the ability to efficiently make the rational improvements in crop species that will lead to increased food production, environmentally friendly agricultural practices, new uses for plants, and even totally new plant-based industries. The most efficient way to gain this understanding is by exploiting the scientific and practical advantages of the model organism Arabidopsis thaliana.
The Multinational Coordinated Arabidopsis thaliana Genome Research Project (1990 – 2001)
The Multinational Coordinated Arabidopsis thaliana Genome Research Project was launched as a cooperative international effort in 1990. At that time, a vision was put forth of a sequenced model genome and the progress that could be made using that genome as a resource in the study of plant physiology, biochemistry, growth and development. A document published in that year, entitled “A Long-range Plan for the Multinational Coordinated Arabidopsis thaliana Genome Project” (NSF document 90-80) put forward a set of research goals and priorities that would lead to complete knowledge of the Arabidopsis genome. By all accounts the project has been a great success. All the goals have been accomplished or surpassed, as summarized in the table on the following page.
International Cooperation
The Genome Project provided a platform for a whole new way of approaching scientific problems. It acknowledged from the outset that members of the Arabidopsis research community are all working toward the same goal; it is therefore to the advantage of all to work in a cooperative manner. By setting forth a goal of international coordination and providing for this coordination by the creation of a multinational committee (the Multinational Science Steering Committee), the Genome Project started and developed with a degree of international cooperation that had rarely been seen in the past. Indeed, the Arabidopsis genome sequencing project (Arabidopsis Genome Initiative – AGI) is widely regarded as a positive example of the progress that can be made when a research community works together to achieve a goal.
Looking back on the project thus far, it is clear that it is this spirit of cooperation that has allowed its success. As genome research enters the next phase of functional genomic research, we must take with us the lessons of the first ten years: international cooperation, coordination of efforts, and communication among the involved groups are essential. By minimizing duplication of effort, removing obstacles for sharing of data and biological resources, and emphasizing the clear realization that all groups are working toward the same goal, we can foster a spirit of cooperation that will well serve the community, and eventually the world.
1990 GOALS | 2000 PROGRESS |
|---|---|
Analysis of the Arabidopsis genome | |
| Saturation mutagenesis and development of facile methods for transposon tagging to define as many genes as possible | Over 500,000 lines of chemically-induced and insertion mutants created and made available to research community |
| Creation of cDNA and EST libraries representing different tissues and cell types | Over 150,000 ESTs in GenBank; cDNA libraries readily available and several full length cDNA sequencing projects underway world-wide |
| Integration of physical and genetic maps | Recombinant Inbred map made available in 1993, used to construct YAC, BAC, and P1 physical maps by 1997 |
| Mapping of centromeres | Completed by tetrad analysis in 1998 |
| Sequence the entire genome | Completion of Arabidopsis genome sequence, December 2000 |
Development of Technologies for Plant Genome Studies | |
| New transformation technologies | Vacuum infiltration and Floral dip technologies, coupled with new-generation transformation plasmids (Agrobacterium T-DNA derivatives) make plant transformation easy and efficient |
| Un-anticipated goal | Expression analysis tools such as DNA chips, microarrays and yeast two hybrid systems became available |
Establishment of Biological Resource Centers | |
| Establish two centers to serve the world-community of Arabidopsis researchers | NASC (http://arabidopsis.org.uk/), ABRC (http://www.arabidopsis.org/abrc/index.html), established in 1991 and together handled over 90,000 orders in 2000 |
Information Sharing and Databases | |
| Development of an informatics program to facilitate exchange of research results | TAIR (http://www.arabidopsis.org), AGR (http://ukcrop.net/agr/), Newsgroups (UK based: Arabuk@lists.bbsrc.ac.uk; US based: arab-gen@net.bio.net) |
Development of Human Resources | |
| Support postdoctoral fellowships for Arabidopsis research | Numerous postdoctoral fellows were trained worldwide under various Arabidopsis research programs. Young scientists dominate the Arabidopsis biology field, indicating successful efforts in human resource development. |
| Support short-term scientist exchanges and short courses for training in Arabidopsis research techniques | Annual Arabidopsis Molecular Biology Course held at Cold Spring Harbor Laboratory |
Workshops and Symposia | |
| Support workshops and symposia to disseminate the results of Arabidopsis research | Establishment and support of an annual International Conference on Arabidopsis Research |
The Genome of Arabidopsis
In December of 2000, the Arabidopsis research community announced a major accomplishment: the completion of the sequence of a flowering plant. For the first time, we have in hand the sequence of all of the genes necessary for a plant to function, knowledge unprecedented in the history of science. Additionally, this sequence is freely available to every member of the scientific community. Below is a summary of major findings described in a groundbreaking paper, “The Arabidopsis Genome Initiative”, Analysis of the genome sequence of the flowering plant Arabidopsis thaliana” (Nature 408, 796-815, 2000).
The genome of Arabidopsis:
- Contains about 125 megabases of sequence
- Encodes approximately 25,500 genes
- Contains a similar number of gene functional classifications as other se- quenced eukaryotic genomes (Drosophila melanogaster and Ceanorhabditis elegans) (see Figure 1)
- Has 35% unique genes
- Has 37.5% genes that exist as members of large gene families (families of 5 or more members)
- Shows evidence of ancient polyploidy: an estimated 58-60% of the Arabidopsis genome exists as large segmental duplications (see Figure 2)
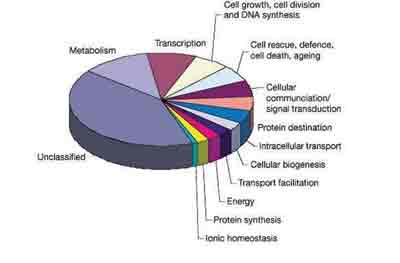
Figure 1: Functional analysis of Arabidopsis genes – Proportion of predicted Arabidopsis genes in different functional categories. Copyright Nature Publications (http://www.nature.com/), as published in Nature 408, 796-815 (14 December 2000). Permission to use kindly granted by Nature Publications.

Figure 2: Summary of segmental duplications in Arabidopsis thaliana. The five chromosomes are depicted as grey horizontal bars, centromeres marked black. Colored bands serve as representations of duplicated segments. Bands are twisted if the corresponding segments have reversed orientation. The scale is in Megabasepairs (Mbp). Courtesy of Dirk Haase of MIPS (http://mips.gsf.de), and copyright Nature Publications (http://www.nature.com/), as published in Nature 408, 796-815 (14 December 2000). Permission to use kindly granted by Nature Publications.
Analysis of the sequence of the Arabidopsis genome tells us that the genome of a higher plant is similar in several important ways to the genome of other sequenced multicellular organisms. It also points out several important differences, which may not be too surprising, considering that plants differ in many important ways from the animals whose genomes have been analyzed. Plants are autotrophic: they require only light, water, air and minerals to survive. They can therefore be expected to have genes that animals do not have, encoding the proteins and enzymes involved in plant-specific processes, including the complex process of photosynthesis.
- Arabidopsis centromeric regions, although largely heterochromatic, overall contain at least 47 expressed genes
- Arabidopsis contains several classes of proteins that are used in animal sys- tems for processes not present in the plant, underscoring the idea that evolution makes use of the tools it is given to accomplish different tasks in different organisms
- Plants have evolved a host of signal transduction apparati, perhaps to enable them to deal with their sessile nature
- Arabidopsis genome contains genes encoding RNA polymerase subunits not seen in other eukaryotic organisms
- Arabidopsis has genes unique to plants – approximately 150 unique protein families were found, including 16 unique families of transcription factors
- Arabidopsis has many gene families common to plants and animals which have been greatly expanded in plants – for instance, Arabidopsis contains 10- fold as many aquaporin (water channel) proteins than any other sequenced organism
The Multinational Coordinated Arabidopsis thaliana Functional Genomics Project
Project Goals
In 1990, a set of research priorities and goals were put forth which would lead to the knowledge of a complete plant genome sequence. With that goal successfully met, the time has come to put forth a new set of goals.
In this respect, probably the most important information provided through the analysis of the Arabidopsis genome sequence was the discovery of the limits of our current understanding of plant gene function and of the roles that the genes play in the multiplicity of processes involved in plant metabolism, development and interaction with the environment. The several decades of pre-genome research in Arabidopsis has yielded experimental data on less than 10% of Arabidopsis genes. It is the task of the Arabidopsis research community to ensure that in the same timely and cooperative manner as we approached the challenge of sequencing the genome, we use the genome information to understand the function of all Arabidopsis genes and in this way to achieve comprehensive knowledge of plant biology.
In the genome sequencing era, the many groups making up the Arabidopsis genomics community were working toward a single goal. Technologies and data converged on the single endpoint of a sequenced genome. The destination of a sequenced genome has now become a launchpad; from this launchpad will spring many technologies and types of data with which we will move forward to the new, multi-pronged goal of complete functional knowledge of an Arabidopsis plant.
The Multinational Coordinated Arabidopsis Functional Genomics Project is an idea that developed from a workshop that was held in early 2000 entitled “Functional Genomics and the Virtual Plant: A blueprint for understanding how plants are built and how to improve them” (text available at http://www.arabidopsis.org/info/workshop2010.html). Resulting from the workshop were new objectives for the Arabidopsis community, “to exploit the revolution in plant genomics by understanding the function of all genes of a reference species within their cellular, organismal and evolutionary context by the year 2010.” The details of this project can be viewed as the second phase of the far-reaching vision described by the scientists who launched the Multinational Coordinated Arabidopsis thaliana Genome Research Project in 1990.
To achieve a complete understanding of the biology of a plant, we must in essence create a wiring diagram of a plant throughout its entire life cycle: from germinating seed to production of the next generation of seeds in mature flowers. These processes are controlled by genes and the proteins they encode. They are directed by both intrinsic developmental cues and environmental signals. The long-term goal for plant biology following complete sequencing of the Arabidopsis genome is to understand every molecular interaction in every cell throughout a plant lifecycle.
The ultimate expression of our goal is nothing short of a virtual plant which one could observe growing on a computer screen, being able to impose environmental changes and to stop this process at any point in that development, and with the click of a computer mouse, accessing all the genetic information expressed in any organ or cell and the molecular processes mediated by these factors.
Complete knowledge of the workings of a plant – a “virtual plant” – will allow a profound understanding of the biochemical processes and physiological responses of a plant. This knowledge will allow hypothesis testing and experimentation leading to the modification and improvement of crops. It will result in a future in which we can limit our dependence on chemical pesticides and fertilizers, lessen our negative impact on the earth, and maximize crop yields to feed a growing world.
Scientific Objectives
The objectives now being put forth for the world-wide Arabidopsis research community include the development of expanded genetic toolkits as a service to the research community, implementation of a whole-systems approach to the identification of gene function from the molecular to evolutionary levels, expansion of the role for bioinformatics, development of human resources, and international collaboration.
A key strength of Arabidopsis as a model is its facile forward genetics, largely due to its relatively small size and short life cycle. One can isolate mutants disrupted in many processes and study the effects of each mutation. Despite this, roughly 40% of the genes found in the genomic sequence do not encode a protein of predictable function.
Thus, out of the 25,500 predicted genes in the Arabidopsis genome, ca. 10,000 have a sequence that tells us nothing about what they do. For instance, sequence reveals that there are about 1,200 protein kinases in the Arabidopsis genome; however, to date, the in situ functions of only about a dozen of these have been found by forward genetics. Similarly, we know the identity of the ligand that binds the putative receptor site in only a handful of the approximately 600 members of the receptor kinase subfamily.
Since forward genetics relies on a phenotype arising from a single gene mutation, it is likely that a large number of genes may not be easily characterized using this approach. In order to identify functions for these genes, we need to develop a more sophisticated genetic toolkit for both forward and reverse genetic screens.
Overexpression of natural or altered proteins can provide insights into families of genes that are collectively essential. A straightforward, albeit laborious, approach that resembles strategic breeding weds reverse genetics and forward genetics. In this approach, the genome sequence is used to locate protein family members. Knockout mutations are located for all the genes in a family and the lines are crossed so that one plant contains knockout alleles of all the members of the gene family, creating a more robust phenotype. A process like this one can lead to increased understanding of the functions of genes that exist in gene families, and is not possible without the entire sequence in hand.
Short-term goals:
- Comprehensive sets of sequence indexed mutants, accessible via database search
- Whole genome mapping procedures
- Facile conditional expression systems for sensitized and saturation screens for rare alleles
Mid-term goals:
- Development of many combinations of large families of Recombinant Inbred Lines and Genetic Substitution Lines to allow facile analysis of natural variation
- Construction of comprehensive sets of defined deletions of linked, duplicated genes
- Development of methods for directed mutations and site specific recombination
- Establishment of libraries (complete collections) of transgenic lines for short term overexpression or repression of gene function
Goals for 2010:
- Plant artificial chromosomes
- Approaches to allow directed combination of multiple genetic modifica- tions (such as strategic breeding)
The post-genome-sequence era allows a shift from single-gene or single-process research to whole-systems approaches to understanding plant biology. Identified as tools for the goal of global understanding of the plant are global analysis of gene expression, global analysis of protein dynamics, metabolite dynamics, global catalogues of molecular interactions, and comparative genomics.
One of the most critical aspects of this project is the enabling technologies that must be developed to achieve the scientific objectives we are putting forth. The proposed research will lead to a new array of fully developed technologies for scientific investigation, particularly in the areas of proteomics and metabolomics. While these technologies will be developed for the purpose of Arabidopsis research, they will naturally spill over into the world of research on economically important plant species and indeed research on all complex systems.
Global analysis of gene expression
An understanding of gene function begins with knowledge of when and where each gene is expressed during the normal development of a plant. Taken together, this information will become a platform from which the concerted action of gene sets in the formation of tissues and organs can be elucidated. Further, examination of the changes in gene expression that occur with environmental changes will illustrate the dynamic nature of gene regulation in plants.
Short-term goals:
- Construct gene-specific DNA probes for expression analysis at high sensi- tivity and wide dynamic range
- Define full-length cDNAs to facilitate annotation of the genome and subse- quent analyses of protein expression
Mid-term goal:
- Determine global mRNA expression profiles at the organ, cellular and sub- cellular levels under a wide variety of precisely defined (by quantitative mea- sures) environmental conditions
Goals for 2010:
- Identify the cis-regulatory sequences of all genes
- Determine the regulatory circuits controlled by each transcription factor in the genome
- Uncover the in planta role of every gene through forward or reverse ge- netic approaches
Global analysis of the plant proteome
The sum of gene expression changes is translated through development into the proteins from which cellular machines are built. Understanding protein dynamics will enable prediction of what machines exist and how they work throughout a plant’s life cycle. This aspect of Arabidopsis research has become especially important, because recent experimentation suggests that RNA changes alone are remarkably poor predictors for final changes in protein levels or enzymatic activity.
Short-term goals:
- Develop facile technology for heterologous expression of all proteins
- Produce antibodies against, or epitope tags on, all deducted proteins
- Catalogue protein profiles at organ, cellular and subcellular levels under a wide variety of environmental conditions
Mid-term goals:
- Achieve a global understanding of post-translational modification
- Define the subsets of genes encoding small peptides or small RNAs and their gene products, which will require the development of novel analytical tools for analysis and modulation of expression
Goals for 2010:
- Identify biochemical functions for every protein
- Determine three-dimensional structures of representative members of every plant-specific protein family
Global analysis of metabolite dynamics
Plant growth and development is dictated, to a large degree, by the uptake, trafficking, storage and use of low molecular weight metabolites. Plant cellular factories produce a bewildering array of secondary metabolites, upon which a large amount of drug and product discovery are based. Understanding metabolite dynamics will result in more efficient use of soil and water based nutrients and will allow rationally designed food and pharmaceutical production in plant factories.
New, sophisticated mass spectrometry techniques provide the Arabidopsis researcher a different type of microscope: one that allows us to monitor potentially thousands or tens of thousands of small molecules at a time. Using these “microscopes”, the mass spectrometers will allow us to see changes at the molecular level that the eye cannot detect at the morphological level. This will be critical for connecting genetic changes with changes in the expression of enzymes and the metabolic pathways they comprise. Much like we collect RNA data using DNA chip technology, we need to expand our phenotypic screens to include newly developed and expensive instruments that open our eyes to the fascinating and complex chemical world of plants.
Mid-term goal:
- Global metabolic profiling at organ, cellular, subcellular levels under a wide variety of precisely defined (by quantitative measures) environmental con- ditions and in many different genetic backgrounds
Goal for 2010:
- Whole systems analysis of the uptake, transport and storage of ions and metabolites
Global catalogues of molecular interactions
The ultimate arbiters of cellular function are the complex protein machines encoded by the mRNA population in each cell at any time during development. Ultimate understanding of the cellular mechanics of a plant requires a catalogue of molecular interactions that occur in each cell of the organism throughout its lifecycle. This ambitious experimental layer incorporates an understanding of the gene expression, protein and metabolite dynamics of the plant.
Goal for 2010:
- Achieve a global description of protein-protein, protein-nucleic acid, protein- metal, and protein-small molecule interactions at organ, cellular, subcellular levels under a wide variety of precisely defined (by quantitative measures) environmental conditions.
Comparative genomics
Completion of the Arabidopsis genome sequence provides significant leverage for future plant genome projects. The reference genome is a platform from which useful comparisons are simplified. We will ultimately be able to predict the evolution of new gene function by comparative genomics. We can glimpse the power of comparative genomics as a tool to understand plant evolution and diversification through the recent strides made in the understanding of plant disease resistance gene structure.
Therefore, as a part of functional genomics work centered on Arabidopsis, work should be done on other species to enable the comparative genomics that will give us the power to apply the knowledge gained from this initiative to crop production.
Short- and Mid-term goal:
- Identify species for survey genomic sequencing based on an expanded definition of phylogenetic nodes
Goals for 2010:
- Survey genomic sequencing and deep EST sampling or sequencing of gene- rich regions from phylogenetic node species
- Define a predictive basis for conservation versus diversification of gene function
- Complete within-species genomic sequence comparisons
- Develop tools for whole genome population biology
Achieving the above goals will require significant investment in and development of bioinformatics tools and databases from which the information required to build the virtual plant will be stored and extracted. A significant effort in this area must be expended in close coordination with the biological aspects of the project.
Ultimately, the database that we envision will provide a common vocabulary, visualization tools, and information retrieval mechanisms that permit integration of all knowledge about an organism into a seamless whole that can be queried from any perspective. Of equal importance for plant biologists, an ideal database will permit scientists to use information about one organism to develop hypotheses about other, less well-studied organisms. Thus, our goal should be to develop facile tools that permit an individual working outside the model species to formulate a query based on the organism of interest, have that query directed to the relevant knowledge for the plant models, and present the information about the models in a way that can be understood by the plant biology community at large.
Database architecture allowing easy integration with other databases will be an essential component of this effort. Divergent types of data (e.g. expression array data and in situ hybridization, but also precise information about experimental setup and growth conditions defined by quantitative measures) will need to be integrated and archived. The ability to generate these datasets will easily outpace the ability to rationally maintain, manage, and extract utility from this data. Hence, there is a critical need to invest in novel data-mining approaches and to also bolster support for current databases.
Ongoing goals:
- Develop new cellular and whole plant visualization tools
- Attract bioinformatics professionals to direct and aid in database creation and maintenance
The Arabidopsis community has developed into an excellent training ground for plant scientists. The changing paradigm of functional genomics will require new types of training to encourage and facilitate lateral, interdisciplinary approaches to problem solving.
Some of the technologies that will be used in the new era of functional genomics research will be beyond the scope of individual labs and some will require sets of biological reagents that are not feasible for individual labs to produce, such as a complete cDNA library and complete protein and metabolite inventories. Instead, a new paradigm will arise in which Genome Technology Centers will serve the research community at large by providing services and by producing new tools using economies of scale. The Centers will be dedicated to the creation of and providing access to genome-wide tools, rather than the application of genome-wide tools to solving specific research problems. The Centers will thereby enable and facilitate the continued participation of individual labs in functional genomic research.
Depending on the status of development and implementation of the technology, such Centers may be financially supported or may operate self-sustained through user fees. In creating genome-wide tools, the Centers must complement and significantly enable investigators throughout the world. Individual investigators will be at once the main clientele for the Centers and, as the experts in specific biological topics, the dispersed creators of knowledge. The value of this project therefore depends on significant support being available for individual research laboratories throughout the plant biology research community to leverage investment in both the Arabidopsis genome sequencing project and the proposed Centers to solve a wide range of specific biological problems.
The structure of Genome Technology Centers, providing services and economies of scale for systems-based data generation, is not consistent with the traditional training of doctoral and post-doctoral researchers, and the traditional output measurement of publications. Therefore, skilled technical assistants and research personnel will be needed. We will also still need traditionally trained doctoral and post-doctoral researchers with skills in plant molecular biology, genetics and biochemistry.
Short to Mid-term goals:
- Support the establishment and maintenance of Genome Technology Cen- ters, including support for technology development and for individual labs seeking the services of the Centers
- Establish summer courses or other intensive specialized workshops, which are an additional effective means for continuing education of established investigators in a rapidly moving field like plant biology and for initiation of non-plant biologists into the world of plant biology
On-going goals:
- Encourage interdisciplinary training that specifically seeks a systems-based approach for both undergraduate and graduate level students
- Support short- and long-term post-doctoral fellowships with a focus on exchange visits
At the outset of the Multinational Coordinated Arabidopsis thaliana Genome Research Project, an ad hoc committee was formed, made up of representatives of countries and programs around the world involved in Arabidopsis research. Among the goals of this committee were forging relationships and fostering communication among the involved groups. After some time had passed, the committee coalesced into the Multinational Science Steering Committee, and it was members of this committee who ensured communication among the Arabidopsis research community at large.
Arabidopsis research has now progressed to a stage in which functional genomics will take the forefront. Participants in this exciting new world of genomic research realized that the same type of communication and coordination provided by the Multinational Science Steering Committee during the first 10 years of the Project will be necessary in the new era. Accordingly, the committee has been renewed under the title Multinational Arabidopsis Steering Committee to coordinate various functional genomics activities world-wide.
The Multinational Arabidopsis Steering Committee (MASC) will be composed of representatives from each country with major Arabidopsis functional genomics efforts or coalition of countries with smaller programs. It is open to any country interested in participating. Selection of MASC representatives is left to the discretion of each country. It will meet once a year in conjunction with the International Conference on Arabidopsis Research. Specific responsibilities of the committee are:
- To coordinate programmatic aspects of the Arabidopsis research world- wide
- To facilitate open communication and free exchange of data, materials and ideas among the Arabidopsis research community
- To monitor and summarize progress of scientific activities of participating laboratories
- To identify needs and opportunities of the Arabidopsis research commu- nity and communicate them to funding agencies of participating nations
- To periodically update and adjust the course of the Project
Short-term goals:
- Appoint a full time coordinator for the Multinational Arabidopsis Steering Committee
- Publish a long-range plan for the Multinational Arabidopsis Functional Genomics Project
- Establish and maintain an internet site devoted to functional genomics efforts and communication among members of the world-wide Arabidopsis research community
On-going goals:
- Continue to foster international collaboration and coordination of the project
- Continue to monitor progress, periodically reassess the status of the project, and adjust the goals as needed
- Publish periodic progress reports
The International Arabidopsis Functional Genomics Community
Arabidopsis researchers world-wide have already embarked on the functional genomics phase. In this section, brief descriptions of various national and transnational projects will be provided. Detailed information about the major projects can be found organized by country and project type at the website being developed by the Multinational Arabidopsis Steering Committee (site can be found at TAIR, http://www.arabidopsis.org/info/2010_projects).
Australia and New Zealand
MASC Contact: Geoffrey Wasteneys (geoffw@rsbs.anu.edu.au)
Australia
Australia has a strong tradition in plant scientific research. Many institutions, including the Plant Industry Division of the Commonwealth Scientific and Industrial Research Organisation (CSIRO), the major Universities and private enterprise are engaged in Arabidopsis Functional Genomics work ranging from individual projects to international collaborations through to major resource development. There is as yet no large-scale coordinated program in Australia. CSIRO’s Division of Plant Industry (http://www.pi.csiro.au/HomePage.htm) funds major programs in Plant Genomics. Otherwise, funding is mainly available through the Australian Research Council’s (ARC’s) Discovery and Linkage Grant Schemes (http://www.arc.gov.au/) and the Grains Research and Development Corporation of Australia (GRDC) (http://www.grdc.com.au/). In addition, many Australian-based Arabidopsis researchers receive funding from international Agbiotech companies or collaborate with overseas colleagues.
Researchers in all Australian States and the Capital Territory now use Arabidopsis functional genomics approaches. Projects are generally highly focused, but increasingly involve international collaborators. Canberra, Australia’s capital city, remains a major node for Arabidopsis research activity. Together, CSIRO’s Division of Plant Industry (http://www.pi.csiro.au/HomePage.htm), the Australian National University (ANU) (http://www.anu.edu.au/), and CAMBIA (http://www.cambia.org.au/), is a formidable unit of fundamental, industrial and application-driven research. Despite the disappointing non-renewal a few years ago of the ANU-based Cooperative Research Centre for Plant Science, and the subsequent closure of Groupe Limagrain’s Australian arm (http://www.limagrain.com), plant science research in Canberra remains very active.
The Australian Centre for Plant Functional Genomics is a major initiative announced in 2001. Established jointly by the ARC and the GRDC, the Centre’s objective is to contribute to ensuring that Australia remains internationally competitive in plant science research. Where this new entity will be located and whether it will incorporate Arabidopsis into its research program remains to be determined.
Funding Bodies in Australia supporting Arabidopsis Functional Genomics Research
- Australian Research Council (http://www.arc.gov.au/)
- CSIRO Plant Industry (http://www.csiro.au)
- Grains Research and Development Corporation (http://www.grdc.com.au/)
- The Australian Centre for Plant Functional Genomics (http://www.arc.gov.au/acpfg/default.htm)
- The Australian Genome Research Facility (AGRF) (http://www.agrf.org.au/)
New Zealand
Increasing numbers of New Zealand plant scientists are incorporating Arabidopsis thaliana into their research, and at least six groups are using functional genomics approaches. Funding is principally available through the Royal Society of New Zealand’s Marsden Fund (http://www.rsnz.govt.nz/funding/marsden_fund/index.php#Marsden) and the New Zealand Foundation for Research, Science and Technology (http://www.frst.govt.nz/).
Canada
MASC Contacts:
Bill Crosby (bcrosby@gene.pbi.nrc.ca)
Peter McCourt (mccourt@botany.utoronto.ca)
Functional genomics projects at the University of Toronto
(Coleman, McCourt, Berleth, Cutler, Goring, Guttman, Christendat, Provart)
The Arabidopsis Research Group (ARG) at the University of Toronto, which includes eight research groups housed out of the Department of Botany, was originally established to provide resources and expertise for the Arabidopsis community in Canada. These programs are jointly funded through the Ontario Genomics Initiative (OGI), Genome Canada, the National Science and Engineering Research Council (NSERC) and by private industry. All resources and data will be made publicly available through various databases and international stock centers. Contacts for each program are listed or the ARG program director Dr. John Coleman can be reached directly at coleman@botany.utoronto.ca. Ongoing programs include:
- Collection and characterization of random insertion GFP enhancer trap lines in Arabidopsis (Thomas Berleth)
- Collection and phenotypic characterization of inducible activation tagged lines in Arabidopsis (Peter McCourt)
- Bioinfomatic analysis of transcript profiling and predictions of protein struc- ture in Arabidopsis (Denish Christendat, Nick Provart)
- Functional genomics of protein localization in Arabidopsis (Sean Cutler)
- Functional genomics of receptor-like kinases in Arabidopsis (Daphne Goring)
Functional Genomics Projects at the University of BC
(Bohlmann, Douglas, Ellis, Haughn, Li)
The functional genomics program at the UBC includes participants from the Biotechnology Laboratory, Department of Botany and the Department of Plant Science, along others. The program has recently received diverse funding input in support of its programs, including CFI, NSERC, OTIP, FRBC, HFSP, Genome BC and Genome Canada. Select program elements are listed below.
- The exploitation of Arabidopsis as a model system for studying development, including metabolism and deposition of compounds of importance to wood fiber production in Conifers and Populus species (Ellis, Douglas, Bohlmann)
- Development of TILLING resources, in collaboration with Dr. Steve Henikoff (Fred Hutchison Cancer Research Centre, Univ. Washington, Seattle) in sup- port of the joint Genome Canada project concerned with Abiotic Stress of Crops (Haughn)
- Molecular biology of SAR response pathways in Arabidopsis (Li)
Functional Genomics Projects at the NRC-PBI
(Crosby, Risseeuw)
The PBI program derives from activity initiated in late 1999, under the auspices of the NRC ‘Genomics in Health and Agriculture Initiative’ (GHI). The program was additionally funded by Genome Canada, the Saskatchewan-Canada Agri-Food Innovation Fund and, more recently, has linked to an NSF 2010 project concerned with the functional genomics of the Ubiquitin-Protein Ligase (E3) families in Arabidopsis. The following is a summary of the structural and functional genomics sub-programs:
- A limited EST program for Brassica napus (40,000 runs)
- Development of a T-DNA disrupted population of Arabidopsis (in collabora- tion with J. Ecker, Salk Inst., La Jolla, CA)
- Development of a genomic amplicon microarray for known and predicted genes of Arabidopsis
- Development of a 2-hybrid ‘map’ for proteins involved in the E3 Ligase func- tions in Arabidopsis
- Bioinformatics program including HPC (cluster) support, gene annotation and data integration tools
- Investigation of the role of ASK genes in SCF function in Arabidopsis (Risseeuw)
- Molecular biology of pathogen response signaling in Arabidopsis (Fobert, Dépres)
Functional Genomics Projects at Agriculture Canada, Saskatoon (AAFC-SRC)
(Lydiate, Parkin)
The Saskatoon Research Center of Agriculture Canada is conducting an active program designed to exploit Arabidopsis model system in support of genomics approaches to Brassica crop development. The program is funded by the Agriculture Canada Genomics Program, and is supplemented by recent support from Genome Canada. Program elements include:
- Genetic, physical and bioinformatics approaches to defining the relationship between the Arabidopsis and Brassica genomes
- Targeted EST programs in Brassica and Arabidopsis, with a biological emphasis on cold adaptation and biotic stress response (fungal pathogenesis; insect herbivory)
- Development of an Arabidopsis activation-tagged T-DNA insert population
- Development of SAGE libraries and tools for analysis of gene expression in Arabidopsis and Brassica crop species
China
MASC Contacts:
JiaYang Li (jyli@genetics.ac.cn)
ZhangCai Yan (yanzc@mail.nsfc.gov.cn)
The National Natural Science Foundation of China (NSFC) has set up a major project to support functional genomics research on Transcriptional Regulators in Arabidopsis. The NSFC maintains a website at http://www.nsfc.gov.cn/english/english.html.
European Union
MASC contact: Bernard Mulligan (bernard.mulligan@cec.eu.int)
Information compiled by Karin van de Sande (kvd1@york.ac.uk)
The European Union has highlighted Functional Genomics approaches, including plant genomics, in previous “Framework” research funding programmes. In the current 5th Framework Programme (1998-2002) (FP5) a wide variety of fundamental and applied plant genomics research is supported under the ‘specific programme’ called ‘Quality of Life and Management of Living Resources’ – each project involves participation of several European countries. The following list illustrates some of the on-going funded research projects involving Arabidopsis (funding more than €40 million):
- EXOTIC (Exon Trapping Insert Consortium)
- NATURAL (Natural variation in Arabidopsis thaliana: resources for functional analysis)
- REGIA (Regulatory Gene Initiative in Arabidopsis)
- ECCO (European cell cycle consortium)
- ASSOCIOPORT (Associomics of membrane proteins…in yeast and Arabidopsis)
- TF-STRESS (Transcription factors controlling plant responses to environ- mental stress conditions
- CONFAB (Controlling Fatty Acid Breakdown in order to produce viable oilseeds with increased yields of novel oils)
- EDEN (Enzyme Discovery in hybrid aspen for fibre Engineering)
- GVE (Growth, Vigour, Environment)
- NONEMA (Making plants resistant to plant parasitic nematodes: no access - no feeding)
- GEMINI (Genetic determination of Maritime pine pulp and paper properties)
- EUROPECTIN (Upgrading sugarbeet pectins by enzymatic modification and molecular farming)
- GMOCARE (New methodologies for assessing the potential of unintended effects in genetically modified food crops)
- PLANET (European Plant genome Database Network)
FP5 also currently supports about 30 young scientists with individual fellowships to carry out research on Arabidopsis.
The EU Quality of Life Programme websitecan be found at: http://www.cordis.lu/life/. For a database of existing and past EU funded projects, please go to: http://www.cordis.lu/en/home.html
Planning of the 6th Framework Programme (2002-2006) is now well underway (see http://europa.eu.int/comm/research/fp6/index_en.html). Opportunities for plant science research will be available, for example, in several of the proposed ‘priority thematic areas’. An invitation to submit expressions of interest for ‘networks of excellence’ and ‘integrated projects’ appropriate for the thematic priority areas of FP6 was recently published by the EU Commission (deadline of 7 June 2002). Further details are available on http://www.cordis.lu/fp6/eoi-instruments
France
MASC Contact: Ian Small (small@evry.inra.fr)
The major source of Arabidopsis Functional Genomics project funding in France is Génoplante (http://www.genoplante.org/), a joint venture between public funding agencies (INRA, CNRS, CIRAD, IRD) and several French Agbiotech companies (Biogemma, Aventis CropScience, Bioplante). Génoplante has joined forces with GABI, a similar German initiative, and several joint projects have recently been funded.
Non-Génoplante Programmes
- A panel of sequenced Arabidopsis thaliana full-length cDNAs
- (http:// www.evry.inra.fr/public/projects/cdna/cdna.html)
- Analysis of genetic variability between Arabidopsis thaliana ecotypes (Con- tacts: David Bouchez, bouchez@versailles.inra.fr and Georges Pelletier, pelletie@versailles.inra.fr)
- AGRIKOLA: Arabidopsis Genomic RNAi Knock-out Line Analysis
(http://www.evry.inra.fr/public/projects/agrikola/agrikola.html)
Génoplante-funded programmes
- FLAGdb/FST, an inventory of flanking sequence tags from the Arabidopsis T- DNA collection from Versailles (http://www.flagdb-genoplante-info.infobiogen.fr/projects/fst/)
- CATMA, complete Arabidopsis thaliana microarray (http://jicbioinfo.bbsrc.ac.uk/CATMA/) (Programme involving several EEC countries and funded by Génoplante in France)
- An Arabidopsis ORFeome (http://www.evry.inra.fr/public/projects/orfeome/ orfeome.html)
- Analysis of the proteome of Arabidopsis (Contacts: Jacques Joyard, jjoyard@cea.fr and Michel Rossignol, rossignol@ensam.inra.fr)
- Metabolomics: several projects are starting that will analyse levels of various metabolites or protein co-factors in Arabidopsis mutants (The Arabidopsis metabolome by NMR and mass spectroscopy, R. Bligny, CEA, Grenoble; Cy tochromes P450, D. Werck, IBMP, Strasbourg; Glycoproteins, V. Gomord, U. de Rouen; Cell wall polysaccharides, H. Höfte, INRA, Versailles)
Génoplante-info database (http://genoplante-info.infobiogen.fr/) will contain data from the following Arabidopsis projects: FLAGdb, the FST database; GENEFARM, a list of fully annotated Arabidopsis genome sequence data with other genomes; and AFPdb, the data produced by the proteome project.
Germany
MASC Contacts:
Thomas Altmann (altmann@mpimp-golm.mpg.de)
Gerd Jürgens (gerd.juergens@uni-tuebingen.de)
Research on Arabidopsis thaliana has a long history in Germany. Many individual German research groups have been using Arabidopsis for analysis of specific topics in plant biology. Furthermore, individual groups and German members of the European sequencing consortia contributed to the analysis of both its genome structure and its sequencing. Functional Arabidopsis genome analysis has recently received strong support in Germany through the implementation of two major research programs supported by the Ministry for Education and Research (BMBF) and the German Research Foundation (DFG). The aims and content of these two programs follow in the paragraphs below.
Genome Analysis in the Plant Biological System (GABI)
http://www.gabi.de/
GABI Coordinator, Jens Freitag (freitag@mpimp-golm.mpg.de)
In 1999, the German plant genome program ‘GABI´ was initiated with the aim of strengthening plant genome research in Germany, establishing a network of competence (to include public and private research groups as well as business companies), enhancing international collaboration, and enhancing the transfer of knowledge into application. GABI is the acronym for “Genome Analysis in the plant biological System.” Financial support for GABI is provided by the German ministry for education and research (about 90% contribution) and private business companies (about 10% contribution). Within the GABI initiative, about half of the 50 million Euro spent over a four-year period has been directed towards supporting work on Arabidopsis thaliana. In GABI, a one-programme-thesis is followed with respect to plant genome research. A fundamental principle of this initiative is to establish a seamless transfer of research results concerning the model organism(s) to real-life application in crop plants. Established rules (http://www.gabi.de/news/notifications/indexe.html) regulate the disclosure and use of research results obtained through GABI activities.
A major aim of GABI is the establishment and support of international co-operations. Developing a linking network of the various national research activities is of particular interest in Europe considering the current splintered structure of European research. A first step towards setting up direct collaborative efforts in Europe is to establish joint research projects between the French plant genome program, Génoplante and the German GABI initiative. Here, the model plant Arabidopsis serves as the front runner and provides a model for international cooperation. The first joint projects will start sometime this year.
Plant technology and resource developments:
- GABI - KAT (Cologne Arabidopsis T-DNA tagged lines) – Contact: Bernd Weishaar (weisshaa@mpiz-koeln.mpg.de)
- GABI - LAPP (GABI Resource Centre: “Large-scale automated plant proteomics”) – http://www.molgen.mpg.de/~gabi/
- The Resource Center and Primary Database in GABI – http://www.rzpd.de/
GABI Bioinformatics centers:
- GABI Primary Database (GABI-PD) – http://gabi.rzpd.de/
- GABI-Info – Werner Mewes (mewes@gsf.de ) and Klaus F.X. Mayer (kmayer@gsf.de )
GABI-funded Arabidopsis research projects: http://mips.gsf.de/proj/gabi/projects/index.html#arabidopsis
Joint Génoplante - GABI projects:
- Sharing sequence data and computer resources on T-DNA transformants of Arabidopsis thaliana – Bernd Weishaar (weisshaa@mpiz-koeln.mpg.de) and Alain Lecharny (Alain.Lecharny@evry.inra.fr)
- Functional genomics of membrane transporters - A European genomics and proteomics resource for plant membrane transporter – Ulf Ingo Flügge (ui.fluegge@uni-koeln.de) and Jacques Joyard (Jjoyard@cea.fr)
- Cell wall: Interactions between components, identification and functional analysis of cell wall proteins and polysaccharide – Martin Steup (msteup@rz.uni-potsdam.de) and Rafael Pont-Lezica (lezica@smcv.ups-tlse.fr)
- Functional genomics of nitrogen utilisation and nitrogen signaling –
Mark Stitt (stitt@mpimp-golm.mpg.de) and Françoise Vedele
(vedele@versailles.inra.fr) - A fifth joint French - German project is in discussion and will come up soon. The working title of this project is: “Evaluation of natural diversity in Arabidopsis accessions for traits of agronomic or basic importance”. The coordinators of this project will be: Georges Pelletier (pelletie@versailles.inra.fr), Mark Stitt (stitt@mpimp-golm.mpg.de) and Thomas Altmann (altmann@mpimp- golm.mpg.de)
Arabidopsis Functional Genomics Network (AFGN)
http://www.uni-frankfurt.de/fb15/botanik/mcb/AFGN/AFGNHome.html
AFGN Coordinator: Lutz Nover (nover@cellbiology.uni-frankfurt.de)
In addition to the GABI program funded by the BMBF, another initiative discussed a year ago has attracted attention in Germany. The Deutsche Forschungsgemeinschaft (DFG) - German Research Foundation – is coordinating with the US National Science Foundation (NSF) to support projects on Arabidopsis functional genome analysis. AFGN, the Arabidopsis Functional Genomics Network, has been established in order to achieve the enormous goal of elucidating the function of all Arabidopsis genes within the next ten years. This will undoubtedly only be possible if additional programmes in other countries also participate.
Information about AFGN-funded projects:
http://www.uni-frankfurt.de/fb15/botanik/mcb/AFGN/Memebers.html
Italy
MASC Contact: Fernando Migliaccio (fernando.migliaccio@mlib.cnr.it)
Groups involved in Arabidopsis functional genomics research in Italy:
Chiurazzi Group (chiurazzi@iigb.cnr.it)
- Investigation into the possibility that the mechanism controlling nodule de- velopment in legumes might be derived from processes common to all plants.
Costantino Group (paolo.costantino@uniroma1.it)
- Dof proteins – Dr. Paola Vittorioso (paola.vittorioso@uniroma1.it)
- Functional genomic analysis of Arabidopsis transcription factors with the REGIA network of the EU Framework 5 – Dr. Paola Vittorioso (paola. vittorioso@uniroma1.it)
- rolD and flower transition – Dr. Maurizio Trovato (maurizio.trovato@uniroma1.it)
Funding from the EU FP5 Programme REGIA project, Ministry of Research grants, and Institut Pasteur -Fondazione Cenci Bolognetti grant.
Marmiroli Group (marmirol@ipruniv.cce.unipr.it
- Isolation of mutants showing resistance to heavy metals from T-DNA tagged collections
Migliaccio Group (fernando.migliaccio@mlib.cnr.it)
- Investigations into the process of gravitropism and auxin physiology
Funding from the European Space Agency (ESA) and the Italian Space Agency (ASI).
Morelli Group (morelli@inn.ingrm.it)
- Functional analysis of the HD-ZIP III family – Dr. Simona Baima (baima@inran.it)
- Functional analysis of the GLABRA2 (HD-ZIP IV family) – Dr. Renato Rodrigues Pousada (pousada@inran.it)
Funding from Ministry of Research grants.
Ruberti Group (ida.ruberti@uniroma.it)
- HD-Zip proteins – Dr. Monica Carabelli (monica.carabelli@uniroma1.it) and Dr. Giovanna Sessa (giovanna.sessa@uniroma1.it)
- Functional genomic analysis of Arabidopsis transcription factors, with the REGIA network of the EU Framework 5 – Dr. Monica Carabelli (monica.carabelli@uniroma1.it) and Dr. Giovanna Sessa (giovanna.sessa@uniroma1.it)
Funding from the EU FP5 Programme REGIA project, the EU FP5 Programme TF-stress project, and Ministry of Research grants.
Soave Group (carlo.soave@unimi.it)
- Study of proteins that interact with H-ATPases proton pumps
- Isolation of Arabidopsis mutants altered in the sensitivity to the photoinhibition
- Isolation of knock-out mutants altered in the active oxygen scavenging pro- cesses, in the sensitivity to UV-B radiation
- Tonelli Group (chiara.tonelli@unimi.it)
- Large-scale Exon-trapping System – Dr. Massimo Galbiati (massimo.galbiati@unimi.it)
- Functional analysis of MYB and NF-Y transcription factors in Arabidopsis – Dr. Katia Petroni (katia.petroni@unimi.it)
Funding from the EU FP5 Programme, Exotic Project, the EU FP5 Programme, REGIA Project, and the Italian Ministry of University and Research.
Japan
MASC Contact: Kazuo Shinozaki (sinozaki@rtc.riken.go.jp)
Arabidopsis Functional Genomics efforts in Japan:
- Collection and phenotype analysis of Ds transposon-tagged lines with the goal of making a library of phenotypes, and collection of full-length cDNAs at RIKEN Genomic Sciences Center (Contacts: Kazuo Shinozaki, sinozaki@rtc.riken.go.jp and Motoaki Seki, mseki@rtc.riken.go.jp)
- The Functional Genomics Research Group of RIKEN Genomic Sciences Center, in collaboration with the Arabidopsis SPP Group of the United States(http://sequence-www.stanford.edu/ara/SPP.html), have made about 8,000 sequenced, full-length cDNAs publicly available through the RIKEN Bioresource Center (Contact: Masatomo Kobayashi, kobayasi@rtc.riken.go.jp)
- Collaborating with the RIKEN group on the phenotypic analysis project is the group of Kiyotaka Okada at the RIKEN Plant Science Center (Contact: Kiyotaka Okada, Kiyo@ok-lab.bot.Kyoto-u.ac.jp)
- Collection of activation tagging lines at RIKEN Genome Science Center (Con- tact: Minami Matsui, minami@postman.riken.go.jp)
- Genome-wide analysis of the cell wall genes (Contact Kazuhiko Nishitani, nishitan@mail.cc.tohoku.ac.jp)
- Analysis of the Homeo-box genes of Arabidopsis and rice (Contact Makoto Matsuoka, makoto@nuagr1.agr.nagoya-u.ac.jp)
- Collection of T-DNA tagged lines created by the Kazusa group, Arabidopsis and Lotus ESTs (Contact Satoshi Tabata, tabata@kazusa.or.jp)
- Distribution and Data accumulation of Arabidopsis cDNA arrays (Contact JCAA, Japan Arabidopsis Array Consortium@isize.egroups.co.jp)
The RIKEN Bioresource Center started in 2002 to collect Arabidopsis resources produced in Japan, such as: full-length cDNAs; Ds tagging lines and Activation tagging lines produced at the RIKEN Genomic Sciences Center; various ecotyptes and mutants from Sendai Arabidopsis Seed stock Center (PI: Prof Mobuharu Goto); and T-DNA tagging lines from Kazusa DNA Research Institute. The PI for the RIKEN Bioresource Center is Masatamo Kobayashi (kobayasi@rtc.riken.go.jp).
For distribution and data accumulation of Arabidopsis cDNA arrays, contact the JCAA (Japan Arabidopsis Array Consortium@isize.egroups.co.jp).
United Kingdom
MASC Contacts:
Ottoline Leyser (hmol1@york.ac.uk)
Karin van de Sande (kvd1@york.ac.uk)
Ian Furner (ijf@mole.bio.cam.ac.uk)
Keith Lindsey (keith.lindsey@durham.ac.uk)
Sean May (arabidopsis@nottingham.ac.uk)
Compiled by Karin van de Sande
GARNet, the Genomic Arabidopsis Resource Network
(http://garnet.arabidopsis.org.uk)
GARNet is establishing the infrastructure and expertise to provide reliable and efficient user-driven and publicly-available functional genomics resources for Arabidopsis research. GARNet started in January 2000 with funding from the UK BBRSC (Biotechnology and Biological Sciences Research Council) for a three year period. The services will move onto a cost recovery basis when BBSRC funding is finished. All GARNet services and resources are publicly available and data created using the GARNet resources will be made publicly available via various databases designed and held at the Nottingham Arabidopsis Stock Centre (AGR, NTP) and the The John Innes Centre (ATIdb).
Services available at GARNet include:
- Transcriptome analysis service (using GSTs from CATMA, http://jic- bioinfo.bbsrc.ac.uk/CATMA) – Sean May (arabidopsis@nottingham.ac.uk), GARNet is also taking part in, and funding the UK part of, the CATMA project, aiming at creating a Complete Arabidopsis Transcriptome Micro Array – Jim Benyon (jim.benyon@hri.ac.uk) and Martin Trick (martin.trick@bbsrc.ac.uk)
- Proteome analysis service – Paul Dupree (p.dupree@bioc.cam.ac.uk) and Kathryn Lilley (ksl23@cam.ac.uk)
- Metabolite analysis – Mike Beale (mike.beale@bbrsc.ac.uk)
- Tools for forward and reverse genetics
- dSPm line generation – Jonathan Jones (jonathan.jones@bbsrc.ac.uk)
- SINS (Sequence of INsertion Sites) Database
(http://www.jic.bbsrc.ac.uk/staff/michael-bevan/atis/index.htm) – Mike Bevan (michael.bevan@bbsrc.ac.uk), Jonathan Jones (jonathan.jones@
bbsrc.ac.uk), and Jonathan Clarke (jonathan.clarke@bbsrc.ac.uk) - GeTCID (Gene Transfer Clone Identification and Distribution service) – Ian Bancroft (ian.bancroft@bbsrc.ac.uk)
- Bioinformatics resources include databases for the proteomics, metabolomics and transcriptomics data that are being developed and held at NASC
The BBSRC Exploiting Genomics Initiative
The BBSRC launched a targeted initiative to allow researchers in the UK to assemble consortia to use Functional Genomics approaches to tackle their research priorities. Below are the plant-related projects funded in this programme.
- Functional genomics of shoot meristem dormancy (Leyser, H. M. O., M. Holdsworth, and M. M. Campbell)
- Exploiting genomics to make glycosidic bonds in vitro and metabolic engi- neering in vivo (Bowles, D. J., G. Davies, R. Edwards, B. G. Davis, and H. J. Gilbert)
- Computational approaches to identifying gene regulatory systems in Arabidopsis (May, S. T., M. W. Bevan, and G. C. Cawley)
- Prediction of protein function in plant genomes using data mining (King, R., H. J. Ougham, and S. T. May)
European Union Framework
Scientists in the UK participate in European collaborations through the European Union’s Framework research opportunities, including CONFAB, EDEN, EXOTIC, GVE, NATURAL, NONEMA, PLANET and REGIA (see European Union section for more information).
Databases
AGR: Arabidopsis Genome Resource (http://ukcrop.net/agr/)
UK CropNet: The UK Crop Plant Bioinformatics Network (http://ukcrop.net/)
ATIdb: Arabidopsis Transposon Insertion database (http://stein.cshl.org/~x-pan/atidb/index.html)
Web sites
GARNet: http://garnet.arabidopsis.org.uk/
Plant-GEMs: http://plant-gems.org/
NASC: http://arabidopsis.org.uk/
JIC: http://www.jic.bbsrc.ac.uk/
Newsgroup
Arab-uk, the UK mailing list to discuss anything Arabidopsis (Arabuk@lists.bbsrc.ac.uk)
United States
MASC Contacts:
Mike Sussman (msussman@facstaff.wisc.edu)
Mary Lou Guerinot (Mary.Lou.Guerinot@Dartmouth.edu)
The Arabidopsis research community in the United States is coordinated by the North American Arabidopsis Steering Committee, consisting of 6 elected members who serve two-year terms. Two members rotate off every year. Two members of the Committee represent U.S. on the Multinational Arabidopsis Steering Committee.
The National Science Foundation (NSF) (https://www.nsf.gov) initiated the Arabidopsis 2010 Project in fiscal year 2001. The program’s goal is to determine the function of 25,000 genes in Arabidopsis by the year 2010. The current foci of the Project are to determine the function of a network of genes and to develop research tools and resources that enable the entire research community to participate in the 2010 activities. NSF requires that the 2010 awards be coordinated with similar activities world-wide, that the investigators post publicly the identity of genes under investigation, and that the outcome of the awards (data, information and materials) be made available to the public according to the timetable approved by NSF. Twenty-seven projects were funded under this program (see https://www.nsf.gov/bio/pubs/awards/2010fy01.htm for a list of awards) in 2001. The NSF expects to continue the Arabidopsis 2010 Project for 10 years, although the focus of the Project may change.
In addition to the Arabidopsis 2010 Project, other activities related to Arabidopsis research are supported by various programs at NSF, including individual research projects, workshops/meetings, information resources and informatics tools development, and the biological resource center. NSF award information can be found at https://www.fastlane.nsf.gov/a6/A6
AwardSearch.htm. The U.S. Department of Agriculture (http://www.reeusda.gov/nri/pubs/abstracts/programlinks01.htm), the U.S. Department of Energy (http://www.sc.doe.gov/production/bes/
eb/ebhome.html) and the National Institutes of Health (http://www.nih.gov), especially the National Institutes of General Medical Sciences, support many research projects involving Arabidopsis, although they do not have a funding program specifically targeted to Arabidopsis research (NIH awards can be searched at http://commons.cit.nih.gov/crisp3/
Crisp_Query.Generate_Screen).



